Plots a prior object
# S3 method for class 'prior'
plot(
x,
plot_type = "base",
x_seq = NULL,
xlim = NULL,
x_range_quant = NULL,
n_points = 1000,
n_samples = 10000,
force_samples = FALSE,
transformation = NULL,
transformation_arguments = NULL,
transformation_settings = FALSE,
show_figures = if (individual) -1 else NULL,
individual = FALSE,
rescale_x = FALSE,
par_name = NULL,
...
)Arguments
- x
a prior
- plot_type
whether to use a base plot
"base"or ggplot2"ggplot"for plotting.- x_seq
sequence of x coordinates
- xlim
x plotting range
- x_range_quant
quantile used for automatically obtaining
x_rangeif bothx_rangeandx_seqare unspecified. Defaults to0.005for all but Cauchy, Student-t, Gamma, and Inverse-gamme distributions that use0.010.- n_points
number of equally spaced points in the
x_rangeifx_seqis unspecified- n_samples
number of samples from the prior distribution if the density cannot be obtained analytically (or if samples are forced with
force_samples = TRUE)- force_samples
should prior be sampled instead of obtaining analytic solution whenever possible
- transformation
transformation to be applied to the prior distribution. Either a character specifying one of the prepared transformations:
- lin
linear transformation in form of
a + b*x- tanh
also known as Fisher's z transformation
- exp
exponential transformation
, or a list containing the transformation function
fun, inverse transformation functioninv, and the Jacobian of the transformationjac. See examples for details.- transformation_arguments
a list with named arguments for the
transformation- transformation_settings
boolean indicating whether the settings the
x_seqorx_rangewas specified on the transformed support- show_figures
which figures should be returned in case of multiple plots are generated. Useful when priors for the omega parameter are plotted and
individual = TRUE.- individual
should individual densities be returned (e.g., in case of weightfunction)
- rescale_x
allows to rescale x-axis in case a weightfunction is plotted.
- par_name
a type of parameter for which the prior is specified. Only relevant if the prior corresponds to a mu parameter that needs to be transformed.
- ...
additional arguments
Value
plot.prior returns either NULL or
an object of class 'ggplot' if plot_type is plot_type = "ggplot".
See also
Examples
# create some prior distributions
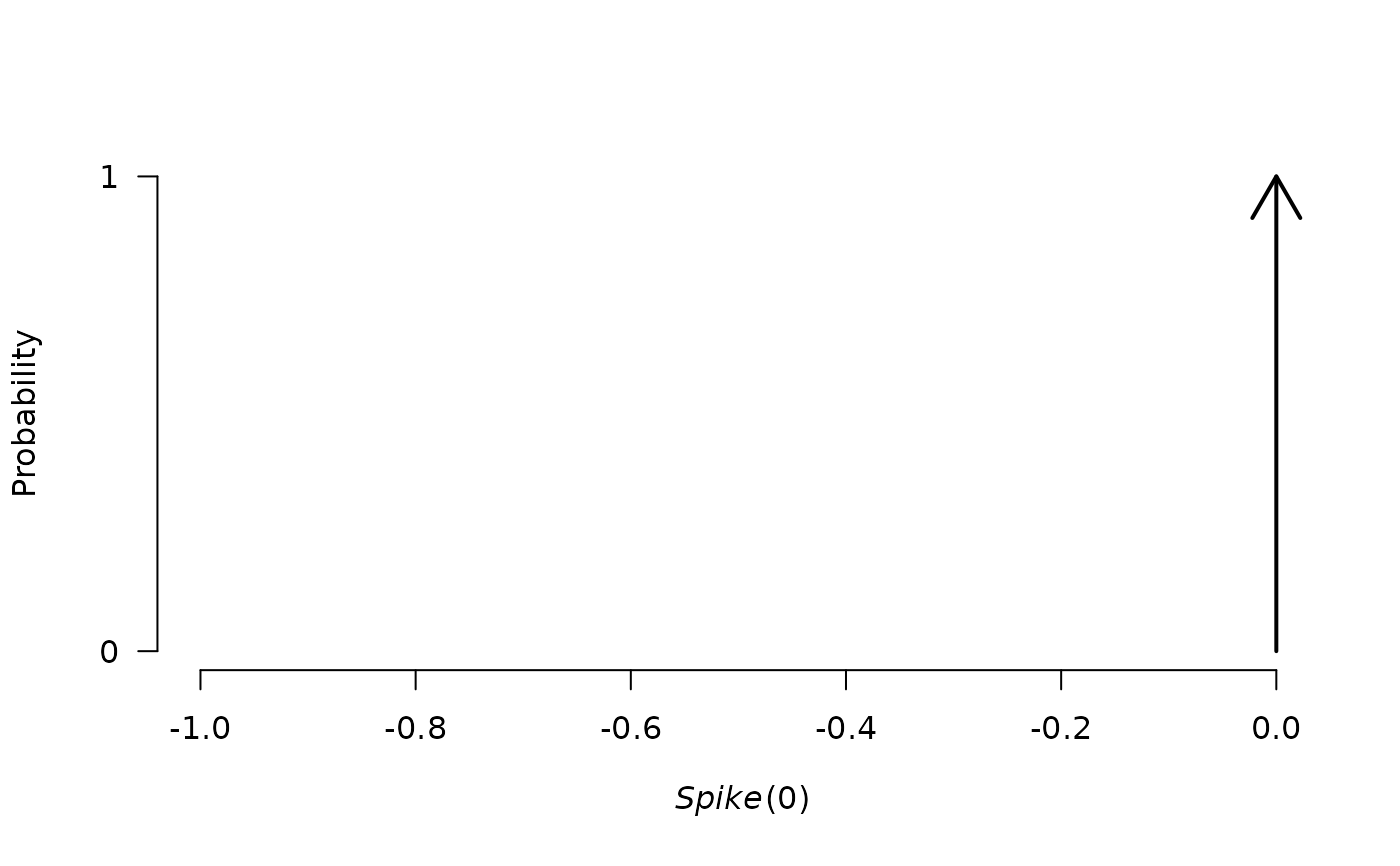
p0 <- prior(distribution = "point", parameters = list(location = 0))
p1 <- prior(distribution = "normal", parameters = list(mean = 0, sd = 1))
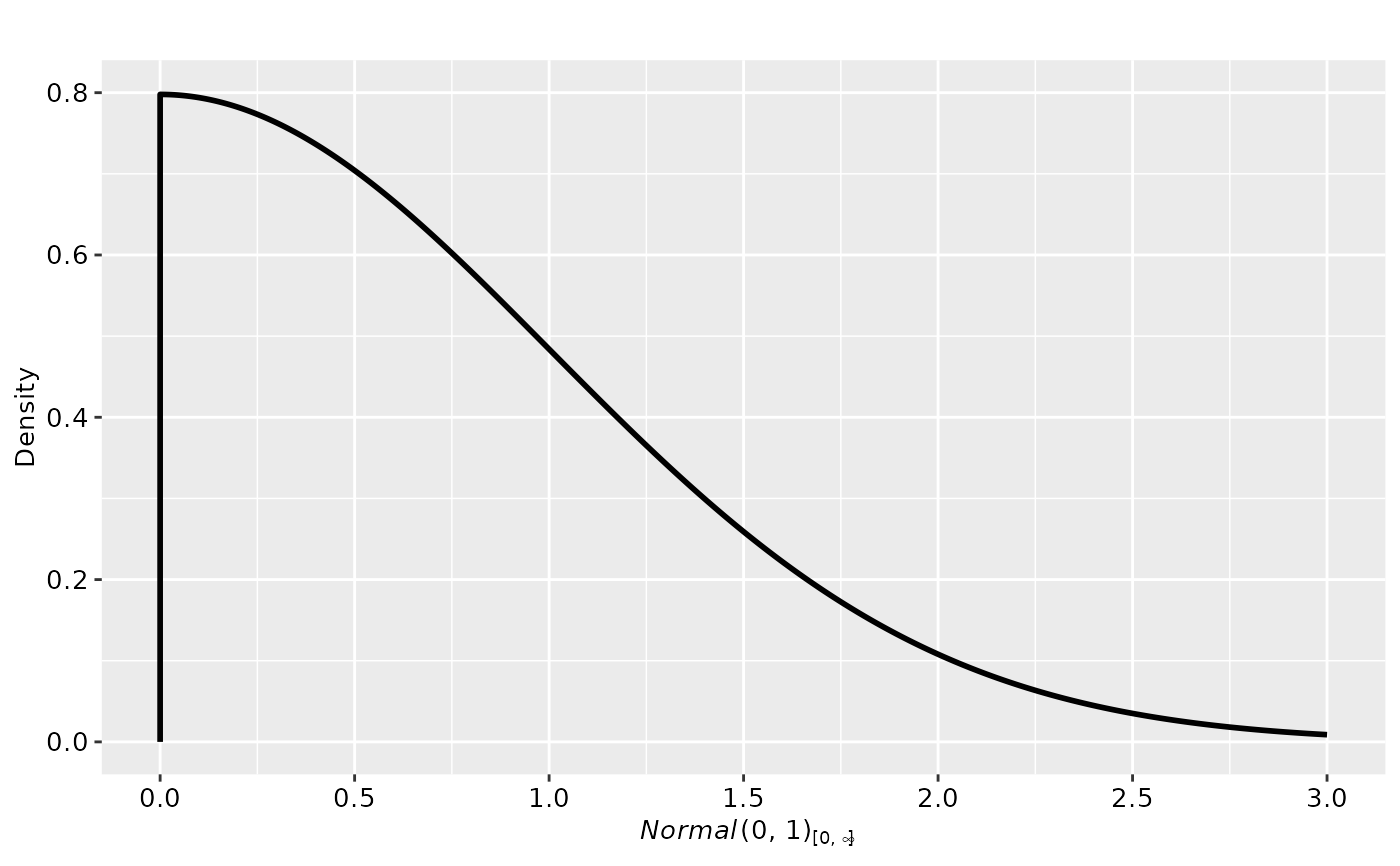
p2 <- prior(distribution = "normal", parameters = list(mean = 0, sd = 1), truncation = list(0, Inf))
# a default plot
plot(p0)
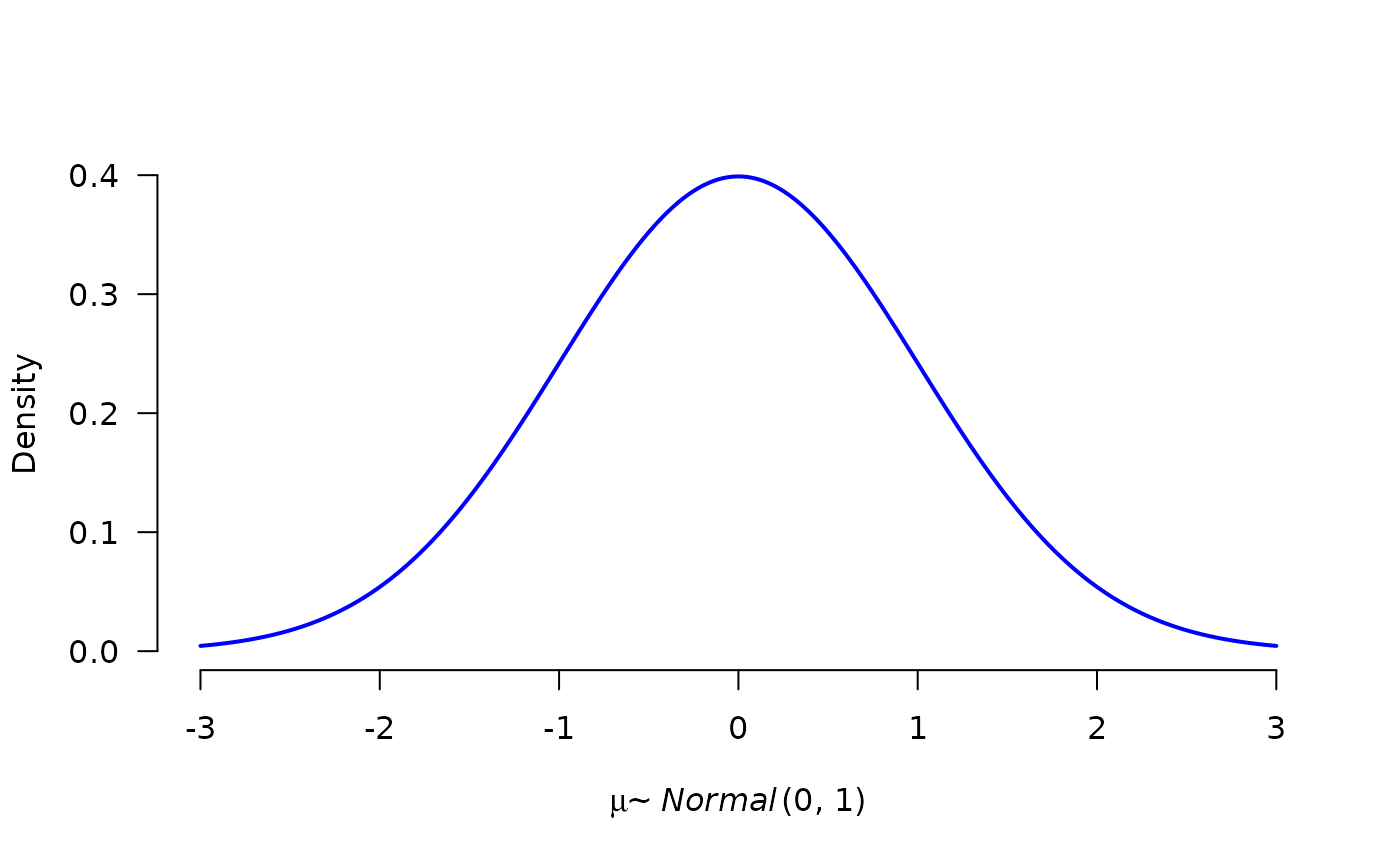
 # manipulate line thickness and color, change the parameter name
plot(p1, lwd = 2, col = "blue", par_name = bquote(mu))
# manipulate line thickness and color, change the parameter name
plot(p1, lwd = 2, col = "blue", par_name = bquote(mu))
 # use ggplot
plot(p2, plot_type = "ggplot")
# use ggplot
plot(p2, plot_type = "ggplot")
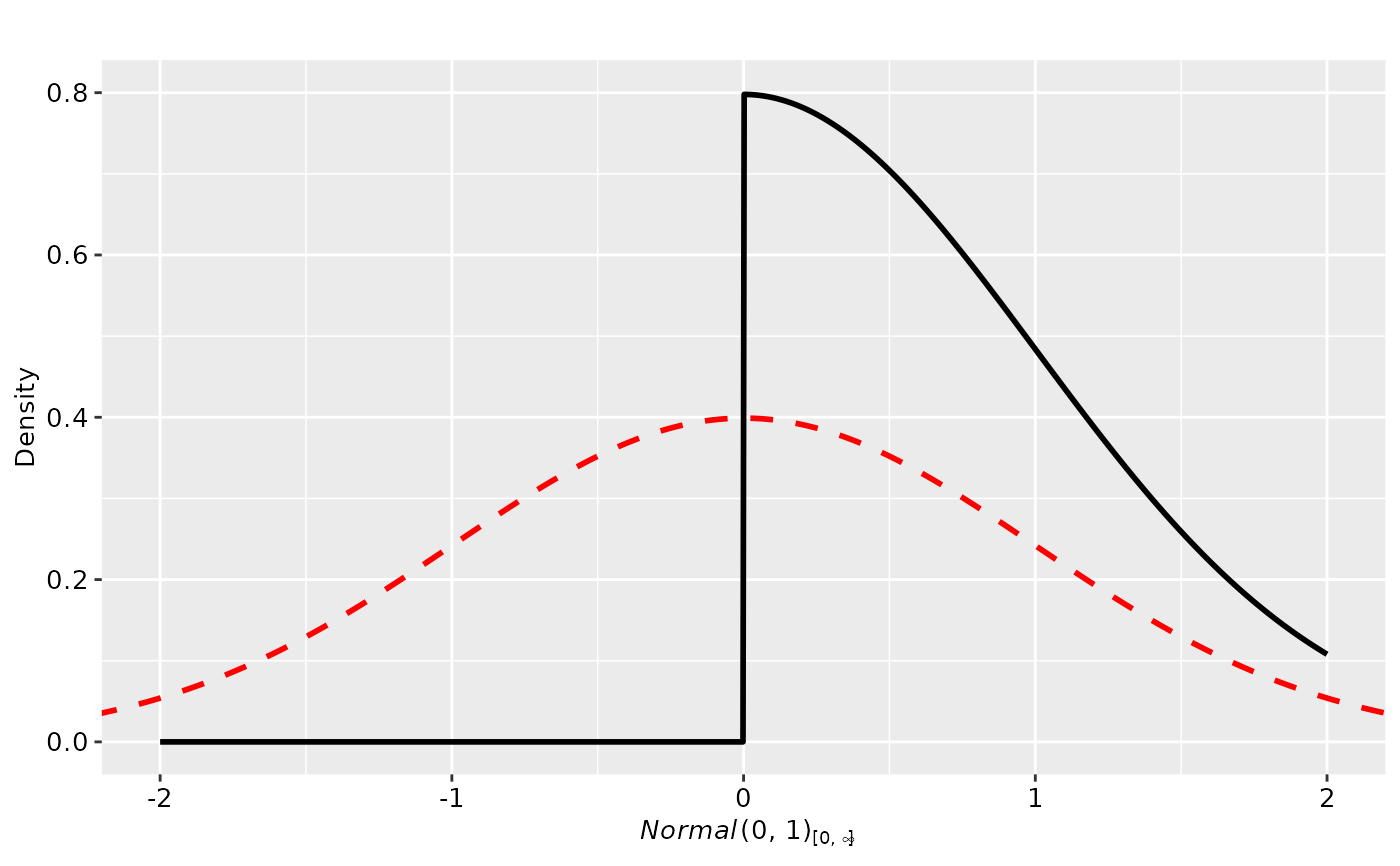
 # utilize the ggplot prior geom
plot(p2, plot_type = "ggplot", xlim = c(-2, 2)) + geom_prior(p1, col = "red", lty = 2)
# utilize the ggplot prior geom
plot(p2, plot_type = "ggplot", xlim = c(-2, 2)) + geom_prior(p1, col = "red", lty = 2)
 # apply transformation
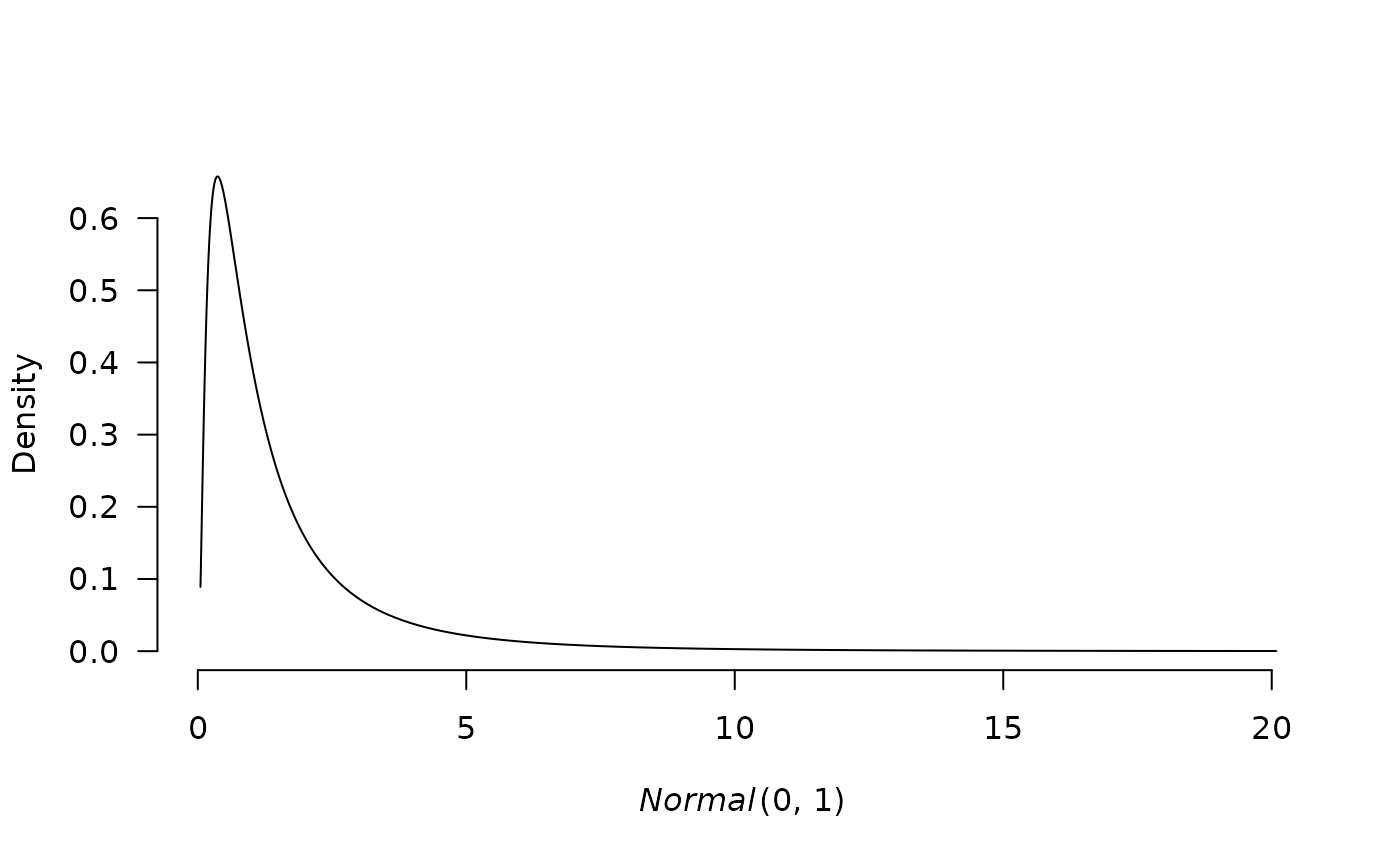
plot(p1, transformation = "exp")
# apply transformation
plot(p1, transformation = "exp")